Why I own Oxford Nanopore Technologies (LON:ONT)
Will the UK finally reap the rewards of its world class genetic sequencing innovation?
Oxford Nanopore is a gene sequencing company meaning that it makes machines which ascertain the sequence of nucleotides (the basic building blocks) in a segment of DNA. There are many valuable applications of gene sequencing including identifying genetic drivers of disease for diagnosis and developing drugs, forensics, understanding evolution, specifying bacteria and viruses, and improving crop and livestock breeding.
The company was founded in 2005 by Hagan Bayley, Spike Willcocks, and Gordon Sanghera. Gordon Sanghera is currently the CEO and Spike Willcocks stepped down from the board last year. Hagan Bayley is currently a professor at Oxford having left the company some years ago.
Illumina - the gene sequencing gorilla
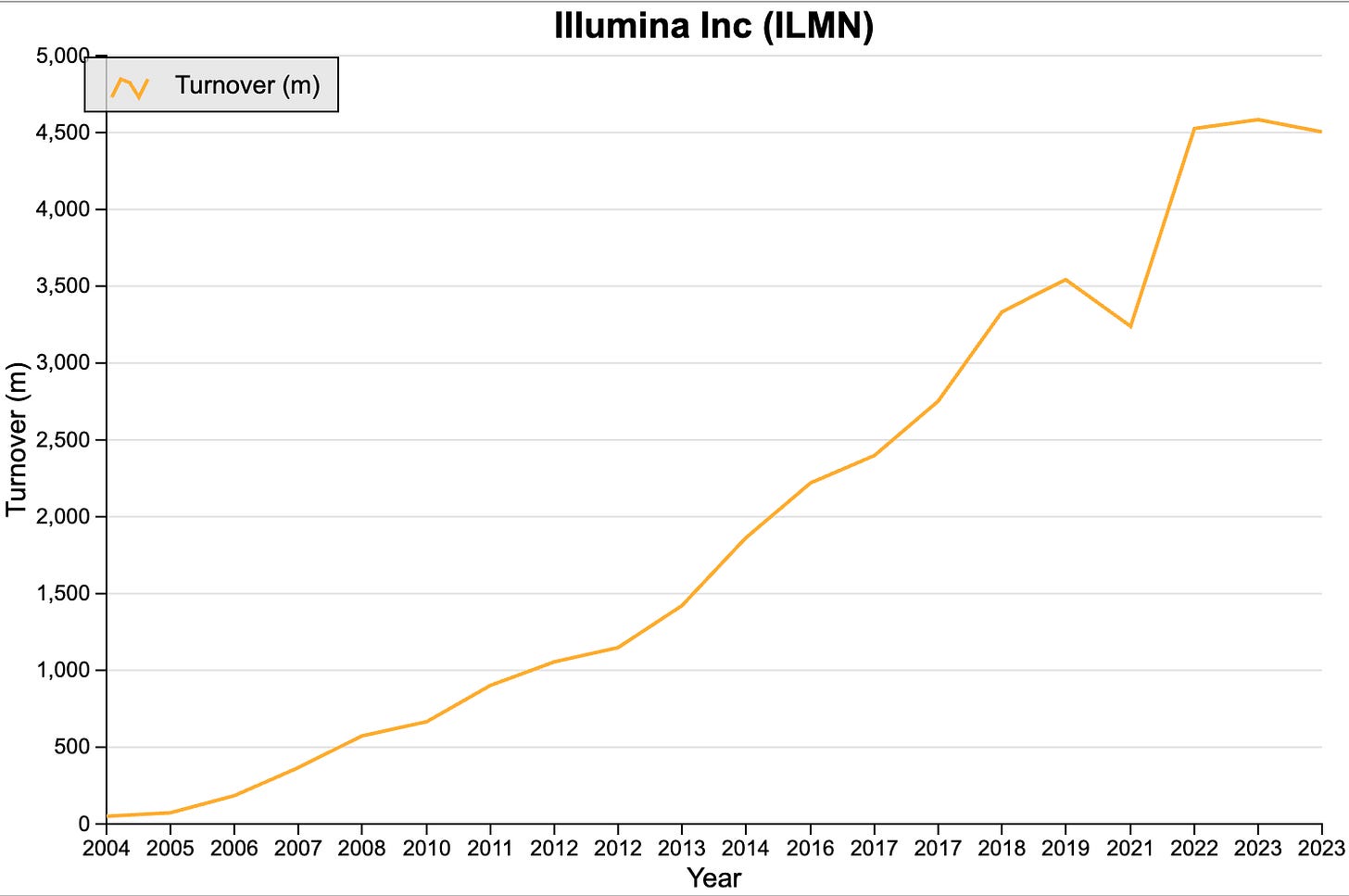
The longtime dominant company in gene sequencing is Illumina (NASDAQ: ILMN). Like Oxford Nanopore, the technology behind Illumina was developed in the UK, but at Cambridge not Oxford. California based Illumina acquired this technology in 2007 and has not looked back since.
Source: SharePad
DeciBio estimates that Illumina has a 90% market share within clinical testing with the remainder held by Thermo Fisher Scientific (NYSE:TMO).
While the clinical sector contributes to more than half of Illumina’s revenue, it is also dominant within the research market. According to Sandiegomics, Illumina’s share of the total sequencing market was around 78% in 2023 with Thermo’s Ion Torrent and Chinese company MGI making up another 14% and Oxford Nanopore at about 5% (70% research).
Illumina offers a range of sequencers of different sizes with prices starting at $50 thousand and going up to $1 million. However, its smaller “benchtop” devices were only introduced late last year and so the vast majority of its business is in large-scale high throughput sequencing performed by big pathology labs. Below is an image of its (somewhat bulky) NovaSeq X system.
Illumina operates a “rozor-razorblade” model whereby it benefits from an installed base of machines which generate additional high margin recurring revenue from ongoing consumables and services. In Illumina’s case, this latter revenue stream constitutes over 80% of total revenue. These consumables include reagent cartridges and library preparation kits.
I’m not going to pretend I know how this stuff works as I do not have a PhD in chemistry. However, all that is important for investing purposes is to realise that once Illumina’s customers commit to a $1 million machine, they are very likely to continue buying its consumables.
This business model explains why it would be difficult for a competitor to displace Illumina from its current customer base, but it does not explain how it ascended to its current position of strength.
The reason Illumina outpaced its rivals in the 2010s is thanks to the technology that Illumina acquired from Cambridge University spinout Solexa. Interestingly, that technology superseded the previous industry standard called Sanger sequencing pioneered by Cambridge academic and two times Nobel prize winner Frederick Sanger. The UK has unrivalled pedigree in this field.
Solexa’s Sequencing-by-Synthesis (SBS) enabled a massive reduction in the cost of sequencing due to its ability to sequence millions of DNA fragments in parallel, whereas Sanger can only sequence one fragment at a time. This breakthrough is the primary reason that DNA sequencing has become so widely adopted today.
That’s enough about Illumina for now, but I’m sure you’re wondering how it is that Oxford Nanopore can be a good investment when Illumina’s market position appears insurmountable. Before tackling that question directly, I have a few comments on Oxford Nanopore’s share register.
ONT’s share register
Source: Stockopedia
Top shareholder, EIT Oxford Holdings (11.1%), is a vehicle associated with Larry Ellison who founded and still owns roughly 40% of another Oxford Nanopore investor, Oracle (3.7%). I note that Oracle is has a strong presence in healthcare data, and that Oxford Nanopore uses Oracle’s cloud infrastructure. When Oracle invested £150 million in Oxford Nanopore at IPO in 2021, Mike Sicilia, Executive Vice President for Oracle Global Industries said,
“Oxford Nanopore’s innovative sequencing technology is unparalleled in the market for its ability to generate rich, accurate genomic data at any scale, from hand-held devices to ultra-high output installations”
Chinese technology giant Tencent (6.6%) needs no introduction, while more recent additions to the register include Biomerieux (6.2%, October 2023) and Novo Holdings (5%, August 2024). These latter two are significant because they represent backing by the biopharma industry.
Biomerieux is a €4 billion revenue In Vitro Diagnostics (meaning test tube tests, IVD) company which has partnered with Oxford Nanopore to develop IVD for the infectious disease market. More on this later.
Meanwhile, Novo Holdings is the investment arm of Danish weight loss goliath Novo Nordisk.
It is quite unusual in my experience to have four large technology companies on the share register of a smaller peer. These sophisticated capital allocators may not be risking much as a proportion of their net worths, but they are likely to be well informed and to perceive substantial potential upside (why else would they be bothered?).
ONT’s point of difference
The key difference between Illumina and Oxford Nanopore is that the former is limited to short read sequencing while the latter specialises in long reads. Short read means that the DNA is broken up into small chunks (100-300 base pairs, bases are the chemical building blocks of DNA ie A, G, C, T) before running them through the sequencer.
Now, understanding all the nuances which distinguish short read from long read is above my pay scale so I asked Grok 3 to summarise the differences as though it was speaking to an (American) child.
Let’s imagine DNA as a super long book that tells the story of who we are—like a giant instruction manual for life. Scientists have two main ways to read this book: short read sequencing and long read sequencing. These methods are like different reading styles, and each one has its own strengths. Here’s what they mean in simple terms and why their differences matter.
Short Read Sequencing: Reading in Tiny Snippets
Think of short read sequencing like flipping through the book and only reading a sentence or two at a time. It’s as if you’re skimming little bits here and there.
What it’s good at: This method is great for spotting small mistakes—like typos in the book. In DNA terms, these typos are tiny changes called SNPs (single nucleotide polymorphisms) or small insertions and deletions (indels). It’s also super helpful if the book is old or torn up (like degraded DNA samples), because you can still read those small pieces.
Why it’s handy: It’s cheaper and fast, so scientists can use it to check lots of books (or DNA samples) without spending too much.
The catch: Since you’re only reading little snippets, it’s tough to figure out how the whole story fits together. Imagine if the book has a line that repeats over and over—like a song chorus. You might not know where each repeat belongs in the story, which can make the big picture confusing.
Long Read Sequencing: Reading Whole Chapters
Now picture long read sequencing as sitting down to read entire chapters of the book at once. You get to follow the story from start to finish without jumping around.
What it’s good at: This method shines when you need to see the big stuff—like if a whole chapter is missing or added (these are called large structural variants in DNA). It’s also awesome for figuring out how different parts of the story connect, especially if some chapters look similar (this is like “phasing haplotypes” in DNA, which helps tell apart the contributions from your mom and dad).
Why it’s handy: You get a clearer view of the book’s plot—how everything links up—which is key for understanding complex DNA stories.
The catch: It’s more expensive and needs a book in good shape (high-quality DNA). Plus, while you’re focused on the big flow, you might miss some tiny typos along the way.
Why Do These Differences Matter?
So, why should you care about these two ways of reading DNA? It depends on what you’re trying to find out:
If you’re hunting for small errors—like a single wrong letter—or working with a beat-up book, short read sequencing is your go-to. It’s affordable and precise for those little details, which might matter for things like finding specific disease-causing mutations.
If you want the big picture—like how the whole story unfolds or spotting huge changes—long read sequencing is better. It’s perfect for understanding complicated DNA layouts, which can be crucial for things like studying how genes are arranged or tackling tricky genetic puzzles.
Sometimes, scientists even team up both methods to get the best of both worlds: the tiny details and the full storyline. Whether it’s catching a small typo or piecing together a sprawling tale, these tools help unlock the secrets in our DNA book—and that’s why their differences are so important!
So a key reason why you might prefer a long read to a short one is that some diseases are not detectable (or fully detectable) using short reads. For example, the structural variants mentioned above are implicated in autism and schizophrenia and so long read might be preferable to short read in these cases.
I asked Grok,
“In what proportion of cases would short reads miss important information in genetic disease?”
Its reply was approximately 10% - 20%.
I am obviously just taking this on face value as I don’t know any better and in any case it may not seem like much. However, remember that long reads also include all the information picked up by short reads. Therefore, if all else were equal, why would you bother going to the expense of developing a test using short read technology for a disease where it is known that short read misses some crucial information in some significant proportion of cases?
Or, as ONT CEO Gordon Sanghera put it,
“Twenty-five percent of some of the hardest diseases (developmental disorders, CNS, neurodegeneration) are all wrapped in the dark genome. Nobody else can get at it at our scale in the way we can.
But over time, as we bring down costs—and we have a road map to halve the cost of a human genome—we will start to see overlap because it will just be natural. You can get everything with long reads that you get from short reads. And, you’ll be able to get it at the same sort of price point with all the additional data that you don’t have today.”
However, this is a mere possibility as we stand today because Illumina remains cheaper at scale and also more accurate.
The diagram below gives an indication of the strides made by Oxford Nanopore to improve accuracy in recent years.
(The Duplex figure of 99.8% is where both strands of DNA are read, one after another which improves accuracy compared to Simplex which involves reading just one strand.)
Source: Oxford Nanopore 2021 Annual Report
Things have improved further since 2021 and in the recent full year 2024 investor presentation, the company states that raw read (Simplex) accuracy is now approaching Q30 (99.9%).
Meanwhile, Illumina is already hitting Q30 accuracy and higher. According to them,
“When sequencing quality reaches Q30, virtually all of the reads will be perfect, with no errors or ambiguities. This is why Q30 is considered a benchmark for quality in next-generation sequencing (NGS).”
So Oxford Nanopore is closing in on Illumina’s benchmark definition, which I assume means it will be good enough for diagnostic purposes.
Pacific Biosciences of California (NASDAQ: PACB) - the long read competition
Pac Bio is Oxford Nanopore’s only long read competitor with about 4% market share in 2023 according to Sandiegomics. Their machines cost $169 thousand for a smaller Vega benchtop model and $779 thousand for a high throughput Revio system.
Remember how Illumina introduced a smaller benchtop range just last year? Well, so did Pac Bio who launched the Vega in November. These are lower throughput machines designed for smaller labs which do not have the workflow or resources to justify shelling out ~$1 million.
I believe that these recent moves to introduce smaller scale machines are a direct response to Oxford Nanopore which has progressed from the other end of the scale.
Its palm sized MinION was the first product introduced in 2015 and costs £2,450. It is ideal for use in the field, where it has no competition.
The low cost and highly accessible nature of MinION helps Oxford Nanopore to “seed the market” as it drives familiarity with ONT’s technology among the hobbyist and academic community.
Awareness among these groups may in time leach into the broader laboratory industry as I imagine that many lab technicians would have passed through a university or been interested in genetics growing up. This might help Oxford Nanopore to chip away at Illumina’s installed base over the longterm.
Oxford Nanopore launched the larger GridION in 2017, followed by the bigger still PromethION in 2018, and finally the largest device, the PromethION 48, in 2019. PromethION 48 is the most expensive and highest throughput device costing £360 thousand.
While Oxford Nanopore’s range of systems is generally cheaper than competitors Illumina and Pac Bio, they are lower throughput and ultimately what counts is cost total cost per genome.
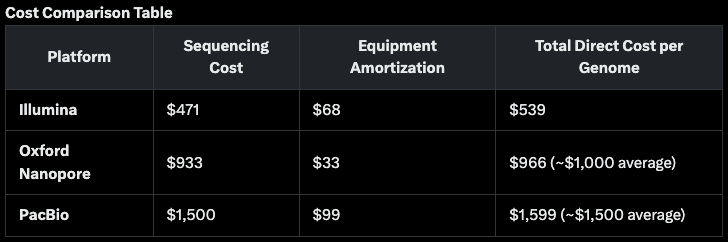
Here is what Grok came back with when I asked it for a cost analysis of the respective companies.
This is above Oxford Nanopore’s claim of $690 today, falling to $235 in the future.
Source: Oxford Nanpore 2024 results presentation
Illumina claims it can deliver a $200 human genome already and Pac Bio reckons it can do the job for under $500.
I conclude from this Illumina is the cost leader and Oxford Nanopore and Pac Bio are trailing, but chasing hard. This is great news for the industry as it should broaden adoption in an increasing number of use cases.
It is worth returning to the idea that Illumina and Pac Bio might be responding to Oxford Nanopore’s positioning at the lower throughput end of the market with their recent product releases.
Once again, Gordon Sanghera presents a possible explanation,
“DNA sequencing, or the access of our source code today, is somewhat akin to mainframe computing in the early 70s, early 80s. And we at Oxford Nanapore have developed a series of platforms that will bring about a much more affordable, accessible, low cost way of accessing DNA information. In the same way as we saw the revolution from mainframe to desktop to handheld computing in your iPhone today.”
I think it is healthy to treat such bold predictions with some scepticism.
However, it is true that the upfront cost of high throughput sequencers is prohibitive to more widespread adoption, for example, in a point of care setting.
It is also true that to achieve cost efficiency, these large systems require batch feeding which makes them inflexible in rapidly changing environments such as during the pandemic.
Oxford Nanopore’s is uniquely able to provide on site real time analysis while all the other platforms require batches to be gathered and processed before running analysis. This makes them uncompetitive in time critical settings or remote locations.
One example where this difference is important is the ongoing collaboration with Biomerieux to develop a test for antibiotic resistant tuberculosis. Here, Oxford Nanopore’s flexible platform is well suited to tackling a problem primarily afflicting lower income countries.
Source: WHO
Another example is a collaboration with Guy’s and St. Thomas’ NHS Trust which led to improve patient outcomes and is being rolled out across 30 NHS trusts. By quickly identifying patients with antibiotic resistant respiratory infections, doctors were able to change medication in a matter of hours rather than days. In addition, the same workflow will be used to identify the emergence of new pathogens to protect the NHS against future pandemics.
Oxford Nanopore’s differentiation is also visible in its financial performance relative to Pac Bio.
Source: Sharepad, average FX rates from GoogleFinance
Note in the chart above how Oxford Nanopore was a beneficiary of covid while Pac Bio was not. Also, ONT’s growth rate has been much higher even when excluding this covid impact.
74% of Oxford Nanopore’s 2024 revenue related to consumables ($172.3 million) compared to just 45% for Pac Bio ($70 million). So as well as growing more quickly, the quality of revenue that Oxford Nanopore generated was superior.
Roche enters
In February Roche provided details of its soon to be released SBX technology, and, much to my chagrin Oxford Nanopore’s stock slipped to a double digit percentage decline on the day.
Certainly, on the surface the prospect of the deep pocketed Roche with its vast diagnostic network as a competitor is an intimidating prospect, but how worried should we really be?
Luckily, I stumbled upon an excellent write up on the possible impact on incumbents by Keith Robison. He is clearly more informed than me and so I am not going to attempt to improve on his analysis on the impact for Oxford Nanopore. He said,
“While in the prior piece I used Oxford Nanopore’s technology as a reference point for explaining SBX, the performance of the platforms is so different that ONT might be the least affected by the new platform - other than constantly having to explain how they are so different. Roche’s instrument certainly won’t fit in your pocket, can’t perform direct RNA sequencing, can’t detect base modifications on native nucleic acids and most certainly won’t be generating 10 kilobase reads, let alone 100 kilobase or megabase sized ones.
One speed, ONT still has a huge edge on time from extracted/amplified DNA to first data. But if you want a lot of data in a hurry, then Roche has an advantage - though technically if money is no object then with a PromethION P24 running 24 flowcells in parallel you could make it an interesting race, as those 24 flowcells could generate a lot of data before the ~5ish hours to start running on SBX would be over.
The one place where SBX might crimp ONT - besides just sucking up attention and laboratory budgets - is in what I’m calling midi read sequencing. For things like whole transcriptome sequencing, ribotyping and some other applications, SBX’s ability to get to 1200 bases may be good enough for many users. As I’ve mentioned multiple times here, there will be a piece outlining the midi read concept tomorrow (I can actually claim that, as it will be in the can before I post this!).”
So apart from medium length reads, there doesn’t seem to be much overlap.
Keith also touches upon Oxford Nanopore’s ability to detect base modification.
Like Pac Bio, Oxford Nanopore reads native DNA whereas all other technologies convert the DNA into something else prior to sequencing. Reading native DNA enables the detection of base (or epigentic) modifications which cannot be done directly with other approaches.
Methylation is a type of base modification that is strongly linked to cancer.
Although Pac Bio reads native DNA, its ability to detect methylation requires additional computational analysis and high coverage (sequencing the same DNA multiple times).
Therefore, Oxford Nanopore is once again unique in being able to process methylation data in real time with no further processing required. In other words, unlike for the competition, a standard workflow using Oxford Nanopore automatically captures methylation data.
The reason that detecting methylation might be useful is that it occurs in cancer cells prior to other genetic mutations. This opens up the possibility of earlier detection through testing which improves outcomes. Understanding methylation also helps with categorising tumours, predicting treatment response and perhaps developing therapies.
Speaking of categorising tumours, a fascinating recent study used Oxford Nanopore sequencing during surgery to determine brain tumour types in 20-40 minutes.
Telling the narrative
I have huge admiration for the people involved in developing Oxford Nanopore’s technology and it seems to this layperson to be a far more elegant solution than presented by competitors.
However, it must be said that some of the communication coming out of the company is laying it on a little thick. For example, the following statement was included in the 2024 results release:
“we are on the cusp of creating the ‘Internet of Living Things’”
There are also regular references to the “disruptive” nature of the platform in the investor materials.
Perhaps such grand claims will be ultimately proven, but right now it seems to me that the competition can do most of the things that Oxford Nanopore can, and in Illumina’s case it can do them more cheaply and accurately.
The one thing that is unique about Oxford Nanopore is its ability to sequence in real time in the field. At a minimum this should assure them an attractive niche even if Gordon Sanghera’s prediction that sequencing will follow the evolution of the computer industry does not come true.
I understand that it is important to be positive to realise a vision, but I also understand why it might be off-putting to invest in a company which makes such lofty proclamations.
Personally, I am willing to accept a bit of exaggeration here because of the massive progress that the Oxford Nanopore team has made historically. See the improvement in accuracy from 95% at the end of 2018 to approaching 99.9% today. Or the decline in the cost of sequencing a human genome which now positions them within reaching distance of Illumina.
In summary, I think Oxford Nanopore is a differentiated business, but am unconvinced it is truly disruptive at this stage. But that is good enough.
Some cold numbers
Source: SharePad
The Oxford Nanopore share price is currently plumbing all time lows.
Part of the explanation for this could be that the business was listed in 2021 at an egregious valuation and at the height of covid when revenue was inflated.
Source: SharePad
The most recent fall in share price from c150p to c100p was, in my view, due to a combination of Trumponomics thumping global technology, the Roche announcement and a cut to guidance which accompanied the release of the 2024 results.
The company reiterated its medium term target of 30% annual revenue growth and EBITDA breakeven by 2027, but now expects revenue for 2025 to be 20% - 23% down from >30%.
In reality, this is just more Trumponomics given it reflects an adjustment for the group’s exposure to NIH funding in the US of 10% to 15% of total revenue. Sadly, the NIH is in the process of being gutted due to the anti-science views of the Trump administration.
I think it prudent of management to assume the total loss of NIH related business, but perhaps there is a possibility that the losses won’t be as bad as feared given that for now at least, the court system still appears to be working in the US.
Source: Oxford Nanopore 2024 results presentation
The above chart partly explains why management is confident of continued strong growth in the short term despite a tricky US funding environment. It shows that the company has doubled sales headcount since January 2023 resulting in a drop off in monthly revenue per head. Those new hires are now settling in and becoming more productive with opportunities per head rising significantly - hopefully a leading indicator of improved sales efficiency.
Source: Oxford Nanopore 2024 results presentation
It is also reassuring to see that applied markets drove overall growth in 2024 given that constraints on government budgets are likely to continue to impact the research market.
I haven’t really touched on biopharma and applied materials opportunities in this post because they are currently relatively small parts of the sequencing market (see below).
(Also, this piece is probably long enough for those of you who made it this far.)
Source: Oxford Nanopore 2024 results presentation
The cash situation looks ok with over £400 million in the bank, cash flow breakeven guided for 2028 and cash burn running at about £100 million per anum having likely peaked in H2 2023. That said, I would not be surprised to see the company raise further capital if the opportunity arises.
Source: Oxford Nanopore 2024 results presentation
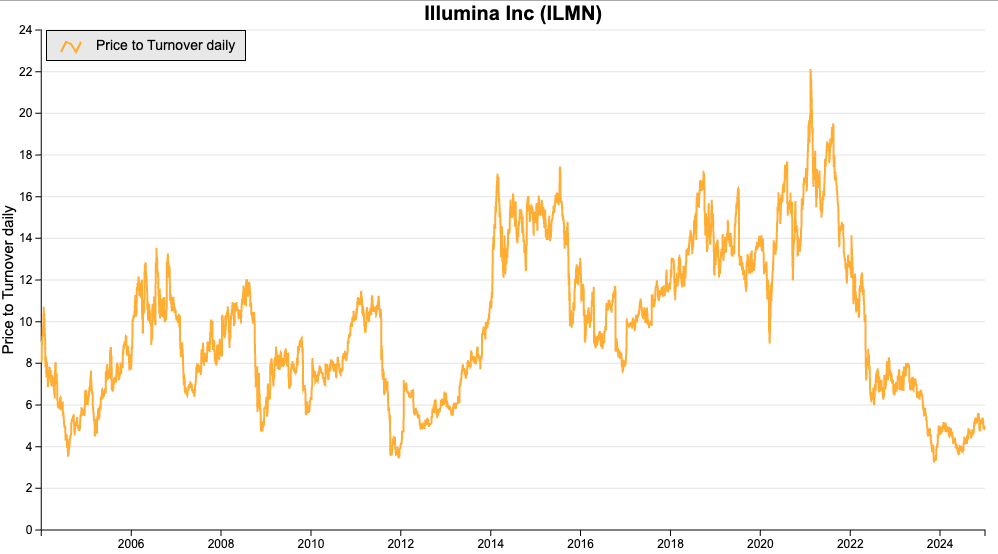
I think a valuation comparison to Illumina is valid because, despite being much smaller, Oxford Nanopore appears to be ploughing a niche and is growing much faster.
Based on the below chart, a price to turnover of eight to ten seems fair compared to Oxford Nanopore’s current rating of five.
Source: SharePad
Consensus revenue forecast for 2027 is £379 million according to SharePad. An eight to ten revenue multiple would equate to a market capitalisation of between £3 billion and £3.8 billion compared to £960 million today.
A final whinge
Oxford Nanopore isn’t perfect, but it is one of the only truly world class technology companies listed in London. I read investors whining all the time that there are no good companies listed in the UK and yet here is one that is totally unloved by the market. You cannot expect to have a thriving stock market if domestic investors do not support it and instead send their money to the US via Vanguard.
Oxford Nanopore is at risk like predecessor Solexa of being gobbled up by a deep pocketed US group. Should that happen, its manufacturing which is currently done entirely in Oxford would be vulnerable to re-shoring. London would lose yet another company from its rapidly shrinking stock market. That would be a shame.